Preliminary DE analysis of P. astreoides translocation
Design
In November 2022, at two sites (MF and CC) located close to each other, coral colonies from 10 m (shallow) and 40 m (deep) were sampled for sequencing (S (shallow) and D (deep)), then each one was split into two pieces. One piece of each colony was put back to the origin depth (SS (shallow -> shallow) and DD (deep -> deep)), and the other was translocated to an opposite depth (SD (shallow -> deep) and DS (deep -> shallow)). All these pieces (SS, DD, SD, DS) were sampled after 6 months, in July 2023, and sequenced.
Overall, the experiment has T0 control (S and D), translocation control (SS and DD) and treatment (SD and DS).
Some of the samples originated from the same location and from a shallow reef (S, SS and SD) are connected between themselves because they came from the same colony. The same connection is present in the samples originated from the same location, deep reef (D, DD and DS).
Goal
The goal is to compare samples between each condition:
- S to D, SS to DD -> shallow and deep conditions depending on a season
- S to SS, D to DD -> controls between each other; seasonal differences between Nov and July
- SS to SD, DD to DS -> the effect of the translocation to an opposite depth
- SS to DS, DD to SD -> how corals are adapting to a depth change
- LRT test to test the effect of depth in general (the difference between full and reduced model)
Sources of variation
- the condition of interest: different conditions (S, D, SS, DD, SD, DS)
- the location (MF and CC)
- intraspecific variation (the same origin of some samples originated from CC S, CC D, MF S and MF D)
- the season (November: S and D; July: SS, DD, SD, DS)
The location and the origin are correlated, and the location is (partly) nested into the origin. The season and the condition correlate as well. The origin also correlates with the condition, since the samples from the same origin depth are connected between each other. I can’t perform a time series design, because the condition is correlated with the time (Nov - July) and I can’t separate effects from each other.
Preliminary PCA
Shallow origin
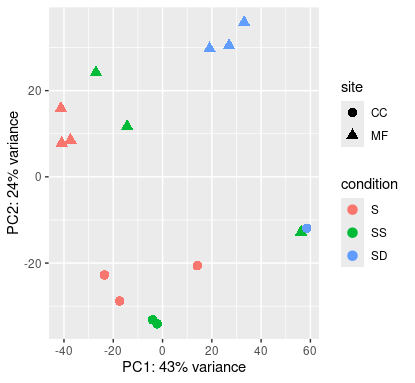
Deep origin
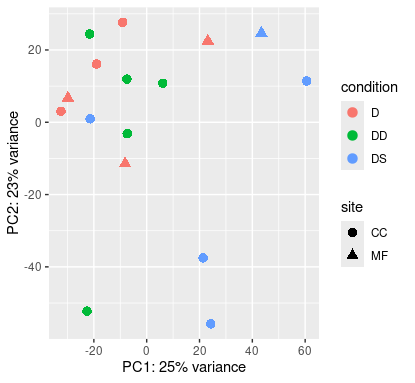
At least at shallow origin, samples are grouped not only by condition, but also by location.
DESeq2 design: shallow and deep origin separately
Metadata tables: shallow origin
If I separate shallow and deep origin, I can use this design:
dds <- DESeqDataSetFromMatrix(countData = countData,
colData = MetaData,
design = ~ origin + condition)
It accounts for intraspecific differences between samples and indirectly for the location, since it is nested into origin.
PCA with manually removed effect (limma::removeBatchEffect) from origin covariate:
shallow
deep
Now, the samples are grouping only by condition. PCA explains more than 90% of variance.
DESeq2 design: all samples together
Since the origin correlates with the condition (there is no connection between samples originated from deep and from shallow reef), I can’t easily include this factor into the model:
Error in checkFullRank(modelMatrix):
the model matrix is not full rank, so the model cannot be fit as specified.
One or more variables or interaction terms in the design formula are linear combinations
of the others and must be removed.
From DESeq2 vignette: this is the case than we want to test the effect of condition while accounting for individual effect:
Finally, there is a case where we can in fact perform inference, but we may need to re-arrange terms to do so.
Consider an experiment with grouped individuals, where we seek to test the group-specific effect of a condition
or treatment, while controlling for individual effects. The individuals are nested within the groups:
an individual can only be in one of the groups, although each individual has one or more observations across condition.
The model with accounting only for the location:
dds <- DESeqDataSetFromMatrix(countData = countData,
colData = MetaData,
design = ~ site + condition)
PCA plot with removed variance from the location:
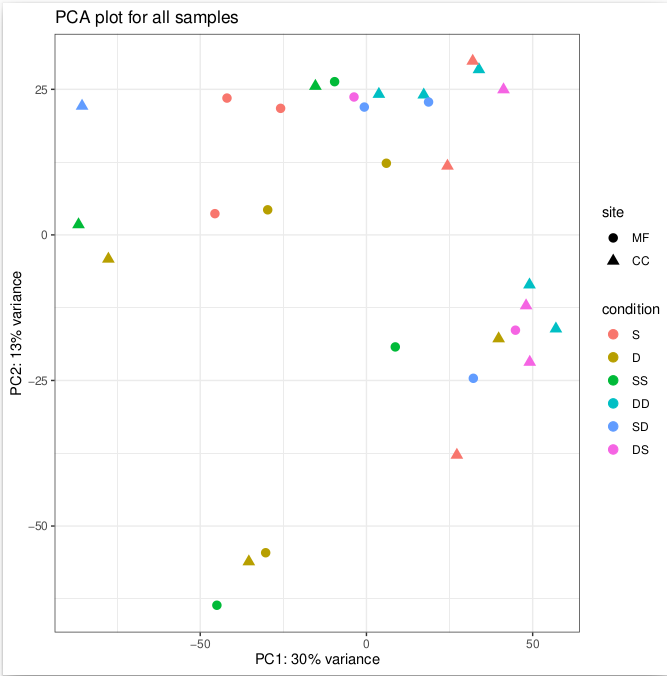
It only explains 40% of variance.